Research
Research in the lab is built on our multi-disciplinary experience in computational bioengineering, cancer systems biology and quantitative pharmacology, and benefits from active collaboration with leaders in clinical oncology, functional genomics, metabolism, and proteomics research. The lab is currently focused on the following areas of research:
1) Single Cell Pharmacology – Multi-factorial analysis of heterogeneous and dynamic cellular responses to perturbations
Our research in this area focuses on the development of new systems biology approaches to study the effect of therapeutic drugs or other forms of perturbation on human cells at a single-cell level. In contrast to classical pharmacology, which relies primarily on population-average measurements to address variance between genetically different cell populations, we aim to understand how clonal populations of cells behave non-identically when exposed to uniform environmental conditions. In this direction, we have combined high-throughput, highly multiplexed, single-cell imaging tools with probabilistic modeling to develop new metrics to evaluate time-dependent drug responses. These metrics reveal not only heterogeneity in drug response, but also its changes with time and drug combinations. Thus, they have important implications for identifying which drugs or drug combinations to explore therapeutically. In addition to measuring the heterogeneity in drug response, we have uncovered its origins at a single-cell level. Specifically, we have investigated transcription factor networks that regulate the expression of genes in response to signaling pathway perturbations and revealed their role in creating the biological noise that leads to population heterogeneity. For example, we have identified the AP-1 family of transcription factors to serve as a central node in linking MAP kinase signal transduction to diverse patterns of cell state plasticity. These studies have advanced our understanding of cellular signaling mechanisms and have important therapeutic implications for heterogeneous diseases such as cancer.
Representative Publications:
Comandante-Lou N*, Baumann DG*, Fallahi-Sichani M. AP-1 transcription factor network explains diverse patterns of cellular plasticity in melanoma cells, Cell Reports (2022). (PubMed Link) (PDF) [* Equal contributions]
Summary
Cellular plasticity associated with fluctuations in transcriptional programs allows individual cells in a tumor to adopt heterogeneous differentiation states and switch phenotype during their adaptive responses to therapies. Despite increasing knowledge of such transcriptional programs, the molecular basis of cellular plasticity remains poorly understood. Here, we combine multiplexed transcriptional and protein measurements at population and single-cell levels with multivariate statistical modeling to show that the state of AP-1 transcription factor network plays a unifying role in explaining diverse patterns of plasticity in melanoma. We find that a regulated balance among AP-1 factors cJUN, JUND, FRA2, FRA1, and cFOS determines the intrinsic diversity of differentiation states and adaptive responses to MAPK inhibitors in melanoma cells. Perturbing this balance through genetic depletion of specific AP-1 proteins, or by MAPK inhibitors, shifts cellular heterogeneity in a predictable fashion. Thus, AP-1 may serve as a critical node for manipulating cellular plasticity with potential therapeutic implications.
Comandante-Lou N, Khaliq M, Venkat D, Manikkam M and Fallahi-Sichani M. Phenotype-Based Probabilistic Analysis of Heterogeneous Responses to Cancer Drugs and Their Combination Efficacy, PLoS Computational Biology (2020). (PubMed Link) (PDF)
Lin JR, Fallahi-Sichani M, Sorger PK. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method, Nature Communications (2015). (PubMed Link) (PDF)
Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW, Sorger PK. Metrics other than potency reveal systematic variation in responses to cancer drugs, Nature Chemical Biology (2013). (PubMed Link) (PDF)
2) Cancer Systems Biology – Adaptive regulation of tumor cell fate
Our research in this area focuses on building a network-level and single-cell understanding of the mechanisms that determine the state of oncogene dependency in tumor cells. Such understanding is key to our ability to predict and ultimately overcome intrinsic and adaptive heterogeneity in responsiveness of tumor cells to therapeutic inhibitors of oncogenic signaling. Among these oncogenes are BRAF-V600E/K/D mutations, which drive tumorigenesis in a wide range of cancers, including >50% of melanomas. The discovery of BRAF oncogenes has led to the development of BRAF/MEK-targeted therapies, but their benefit has been limited in patients due to heterogeneity in the state of BRAF/MEK dependency. To identify the epigenetic regulators of such heterogeneities, we have screened hundreds of epigenetic-modifying compounds across genetically diverse and isogenic populations of melanoma cells. Integrating multiplexed single-cell analysis with multivariate modeling, we have identified three classes of inhibitors that target distinct epigenetic states in melanoma cells. We are currently exploring ways to target these epigenetic states, with the goal of improving therapies for melanoma patients. In another study, we have been studying loss of function of NF1 (NF1-LoF) that drives tumorigenesis in >10% of melanoma patients, for whom currently no effective targeted therapies are clinically available. We combined compound screening and systems biology tools to identify a multi-kinase inhibitor (MTX-216) that is effective in blocking NF1-LoF melanoma cells in vitro and in vivo. Using a combination of kinome selectivity assay, transcriptomic analysis, and genetic experiments, we found the anti-tumor efficacy of MTX-216 to be dependent on its ability to inhibit not only PI3K (its nominal target) but also SYK kinase, and suppression of a group of genes that regulate mitochondrial electron transport chain whose expression is associated with poor survival in NF1-LoF melanoma patients. These studies provide a path to exploit SYK dependency to selectively target NF1-LoF melanoma cells.
Representative Publications:
Khaliq M, Manikkam M, Martinez ED, and Fallahi-Sichani M. Epigenetic modulation reveals differentiation state specificity of oncogene addiction, Nature Communications (2021). (PubMed Link) (PDF)
Summary
Hyperactivation of the MAPK signaling pathway motivates the clinical use of MAPK inhibitors for BRAF-mutant melanomas. Heterogeneity in differentiation state due to epigenetic plasticity, however, results in cell-to-cell variability in the state of MAPK dependency, diminishing the efficacy of MAPK inhibitors. To identify key regulators of such variability, we screen 276 epigenetic-modifying compounds, individually or combined with MAPK inhibitors, across genetically diverse and isogenic populations of melanoma cells. Following single-cell analysis and multivariate modeling, we identify three classes of epigenetic inhibitors that target distinct epigenetic states associated with either one of the lysine-specific histone demethylases Kdm1a or Kdm4b, or BET bromodomain proteins. While melanocytes remain insensitive, the anti-tumor efficacy of each inhibitor is predicted based on melanoma cells' differentiation state and MAPK activity. Our systems pharmacology approach highlights a path toward identifying actionable epigenetic factors that extend the BRAF oncogene addiction paradigm on the basis of tumor cell differentiation state.
Abecunas C, Whitehead CE, Ziemke EK, Baumann DG, Frankowski-McGregor CL, Sebolt-Leopold JS, Fallahi-Sichani M. Loss of NF1 in melanoma confers sensitivity to SYK kinase inhibition, Cancer Research (2023). (PubMed Link) (PDF)
Khaliq M and Fallahi-Sichani M. Epigenetic Mechanisms of Escape from BRAF Oncogene Dependency, Cancers (2019). (PubMed Link) (PDF)
Fallahi-Sichani M#, Becker V, Izar B, Baker GJ, Lin JR, Boswell SA, Shah P, Rotem A, Garraway LA, Sorger PK#. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly-dividing de-differentiated state, Molecular Systems Biology (2017). (PubMed Link) (PDF)
(#Co-corresponding authors)
3) Development of versatile, high-throughput procedures to profile cellular states and their responses to perturbation (with a major focus on multiplexed single-cell imaging)
Advance in biomedical research increasingly depends on the development and application of tools to profile, model and analyze biological systems. Such tools enable us to dissect the interactions between the measured components (e.g., in cells and tissues) and determine how they function together. We have contributed to the development of high-throughput, antibody-based imaging tools for multiplexed profiling of a variety of proteins and their post-translational modifications (representing diverse cellular signaling, metabolic and phenotypic states) across many biological samples at population and single-cell levels. We have utilized these tools to profile cellular responses to perturbations and mechanisms of cell state plasticity in cancer research studies or in other collaborative projects.
Representative Publications:
Hsu J*, Nguyen KT*, Bujnowska M, Janes KA, Fallahi-Sichani M. Protocol for iterative indirect immunofluorescence imaging in cultured cells, tissue sections, and metaphase chromosome spreads, STAR Protocols (2024). (PubMed Link) (PDF) [* Equal contributions]
Summary
Highly-multiplexed imaging technologies have enhanced our ability to study biology at cellular and subcellular resolutions. Among these methods, iterative indirect immunofluorescence imaging (4i) enables generation of multiplexed data on proteins, their posttranslational modifications, and their spatial context within samples via iterations of indirect immunostaining, imaging, and antibody elution. Here, we report streamlined 4i protocols for three sample types. First, we describe a detailed 4i protocol for cultured cells (cell culture-4i), which we recently applied to elucidate the heterogeneity in single-cell abundance of various proteins, including transcription factors, cell signaling, proliferation and differentiation state markers across genetically diverse melanoma cell lines. Then, we describe a 4i protocol for formalin-fixed paraffin embedded (FFPE) tissue sections (tissue-4i), which enables spatial mapping of protein localization at single-cell resolution from whole-tissue sections. Finally, we report a 4i protocol for metaphase chromosome spreads (chromosome spread-4i), which enables mapping of chromosomal proteins at single-chromosome resolution.
Lin JR, Fallahi-Sichani M, Chen JY, Sorger PK. Cyclic immunofluorescence (CycIF), a highly multiplexed method for single-cell imaging, Current Protocols in Chemical Biology (2016). (PubMed Link) (PDF)
Lin JR, Fallahi-Sichani M, Sorger PK. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method, Nature Communications (2015). (PubMed Link) (PDF)
Moerke N, Fallahi-Sichani M. Reverse phase protein arrays for compound profiling, Current Protocols in Chemical Biology (2016). (PubMed Link) (PDF)
4) Computational Biology – Multi-scale and multi-variate modeling of bio-molecular networks
Across a variety of research projects, we have built, refined, and used data-driven, system-wide computational models of cellular function with mechanistic information on the activities of individual biomolecules. We have utilized a diverse range of modeling techniques to uncover important determinants of regulation from complex high-dimensional data spaces and identify measurements of network activity that are predictive of specific cell phenotypes.
Representative Publications:
Summary
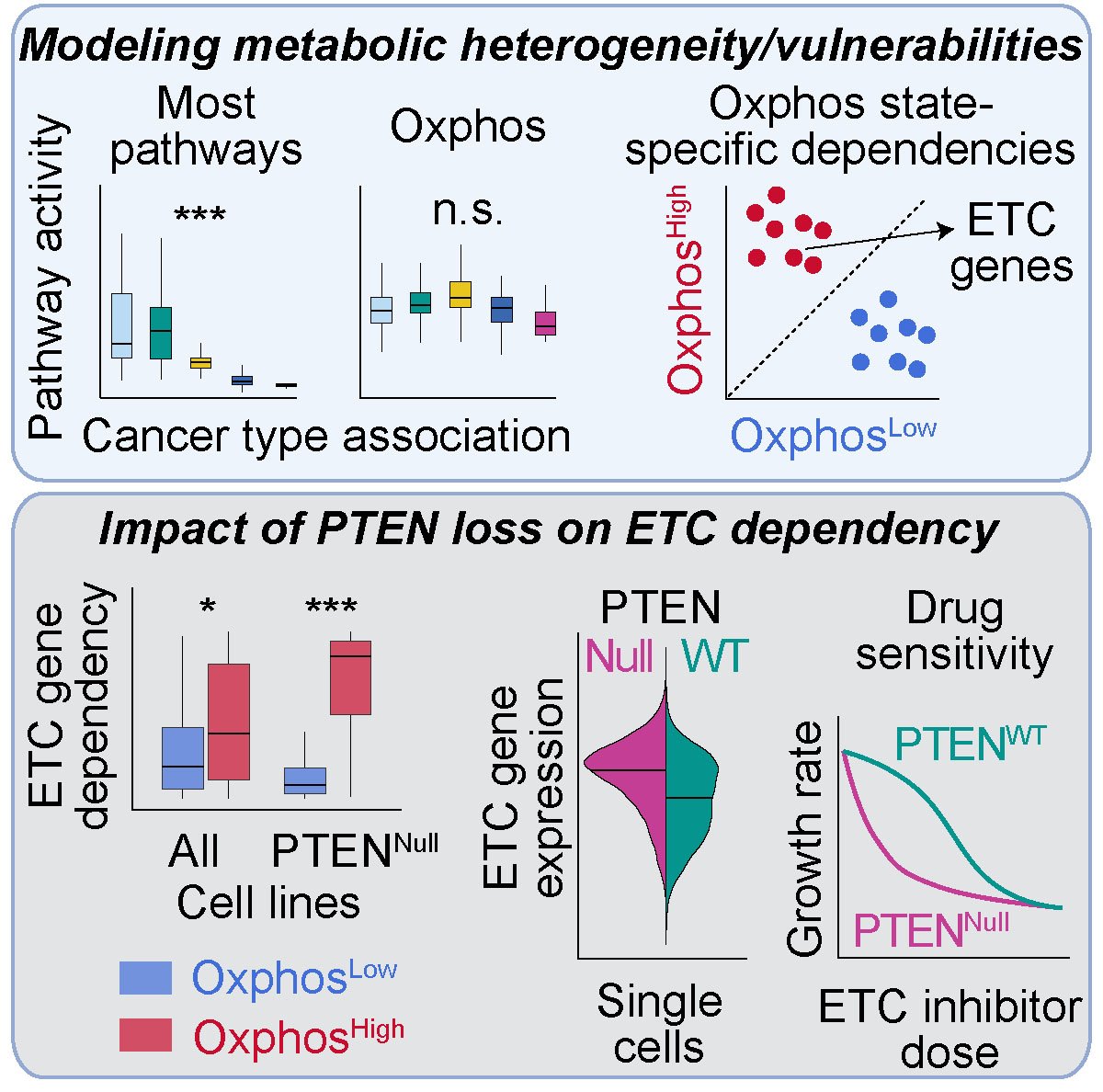
Targeting the distinct metabolic needs of tumor cells has recently emerged as a promising strategy for cancer therapy. The heterogeneous, context-dependent nature of cancer cell metabolism, however, poses challenges to identifying effective therapeutic interventions. Here, we utilize various unsupervised and supervised multivariate modeling approaches to systematically pinpoint recurrent metabolic states within hundreds of cancer cell lines, elucidate their association with tumor lineage and growth environments, and uncover vulnerabilities linked to their metabolic states across diverse genetic and tissue contexts. We validate key findings via analysis of data from patient-derived tumors and pharmacological screens and by performing genetic and pharmacological experiments. Our analysis uncovers synthetically lethal associations between the tumor metabolic state (e.g., oxidative phosphorylation), driver mutations (e.g., loss of tumor suppressor PTEN), and actionable biological targets (e.g., mitochondrial electron transport chain). Investigating the mechanisms underlying these relationships can inform the development of more precise and context-specific, metabolism-targeted cancer therapies.
Comandante-Lou N and Fallahi-Sichani M. Models of Cancer Drug Discovery and Response to Therapy, In Wolkenhauer O (Ed.), Systems Medicine: Integrative, Qualitative and Computational Approaches, vol. 3, pp. 269-276. Oxford: Elsevier (2021). (Link)
Fallahi-Sichani M, Moerke NJ, Niepel M, Zhang T, Gray NS, Sorger PK. Systematic analysis of BRAF(V600E) melanomas reveals a role for JNK/c-Jun pathway in adaptive resistance to drug-induced apoptosis, Molecular Systems Biology (2015). (PubMed Link) (PDF)
Kirschner DE, Hunt CA, Marino S, Fallahi-Sichani M, Linderman JJ. Tuneable resolution as a systems biology approach for multi-scale, multi-compartment computational models, WIREs Systems Biology and Medicine (2014). (PubMed Link) (PDF)
Our research through the years has been made possible with funding support from the following organizations: